The default display regions shown cover the entire sequence if its length is no more than 10,000 nucleotides (nts), and the first 10,000 nucleotides otherwise. The user can change the display position from menu Display->Position, which they can type the regions position (e.g. 0) to position (e.g. 94829). The protein coding genes (or other specific annotated regions) will be displayed as colored blocks with varying sizes (the colors are just used to distinguish one from the next). As you move your mouse, annotation information should pop up both in the display and at the bottom. Click on a gene to select it so you can compare its information to another via highlighting.
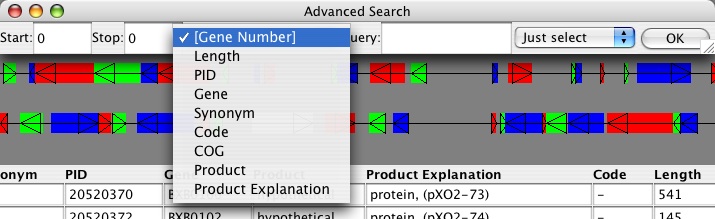
You can also search the annotations for a variety of other criteria, which are dynamically added as the attributes listed by each gene. In this example, you should be able to search based on location (always), Synonym, PID, Gene, Product, Product Explanation, Code, Length, and COG. You can choose to simply select the gene, locate it preserving the zoom (center on it), or re-zoom to it (changes the range to start and stop entered).
To perform a RNA LSS analysis, you need to specify a region to analysis, usually less than 10,000 nts. You can either define the region by menu Display->Position. Or you can click and drag to some smaller section -- say, click near the upper-left corner and drag to past the second gene shown.
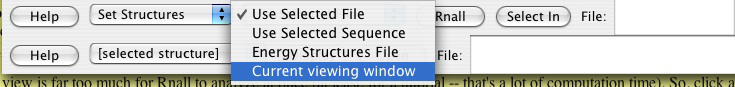
For a thousand base pairs, it will only take a few seconds on a modern fast computer (e.g. Dell Optiplex GX620, Pentium 4 CPU 3.4 GHz, 1G RAM) to predict the secondary structures and display them. Go down to the Rnall control section and make sure Set Structures and Current Viewing Window are selected. The default window size is from 10 to 30, and energy cutoff is 0 Kcal/mol. The users can revise it as they wish.
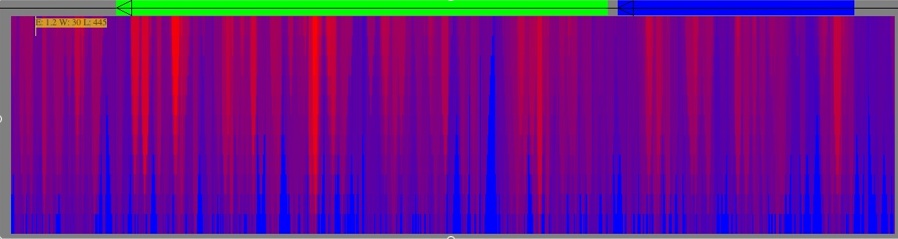
After Rnall calculation, the users will notice now that below the genes colored squares pop up -- these correspond to the structure generated by Rnall. The color represents the energy, its position along the line shows the middle position for the folding, and its height corresponds to the window size used.
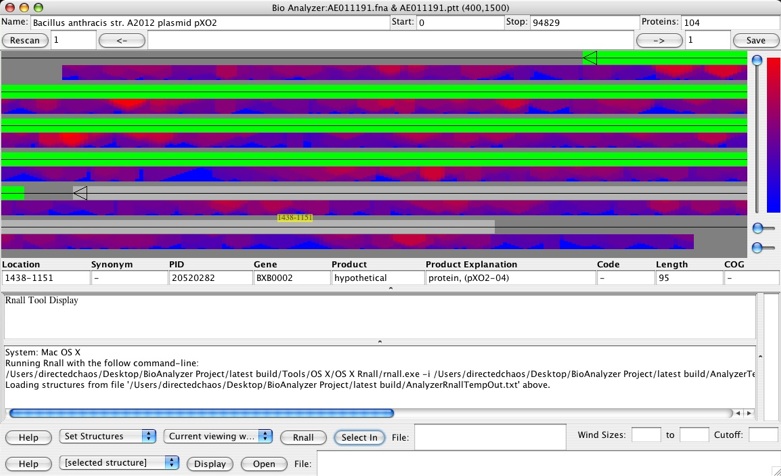
This is not the only way to view it. In the General Preferences you can change the number of rows, and in the Display menu you can quickly switch it between one row and the last number of rows you set in General Preferences. Set it to one row. Now you should see just one long line at the top for the genes, and several rows of energies below.
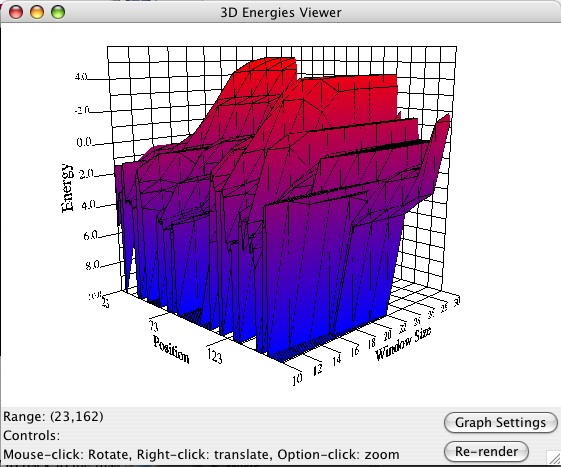
The next part may be more taxing on your system, so zoom in again on a smaller region (the maximum value is 300 nts). Go up to Tools->Energies Viewer and wait a few seconds for it to initialize the 3D representation (you can click cancel on the loading bar to stop and close the window). A new window should pop up -- expand it to however large you want, and transform it with the controls listed. Below is an example:
Go back to the main window. Try clicking on a structure -- it should run some commands in the bottom-left field and give you some information in the top-left field. If GhostScript is working properly and the structure's energy is above the cutoff, it should also display a picture that you can zoom in (single click), zoom out (double click), and scroll (drag).
Another thing you can do in the main window is adjust the colors to focus on a certain energy. Go to Tools->Energies Focuser. Move the left scroll bar down a nudge and then move the right scroll bar down slightly. You should get something like the below:

Experiment. Moving the color bar adjusts the color (energy) it focuses on. Moving the spread bar adjusts the contrast or spread of colors (more down makes it sharper on that specific energy). Finally, the right scroll bar adjusts the over-brightening effect on the target color -- the farther down, the brighter it is. It's not a perfect system, and there are some kinks to work out, but it might help you narrow in on a section quickly (it changes colors by structure, not by pixel). If you move the color bar back to the top it returns the picture to normal.
