CodonO webserver is implemented for synonymous codon usage bias analyses within and across genomes. CodonO measures Synonymous Codon Usage Order (SCUO) in a genomic scale by connecting directly GenBank genomic database. CodonO plots SCUO and GC compositions. The webserver displays the results for multiple genomes in the same plots. It identifies the outliers for a genome or a group of sequences based on Tukey statistical analysis. It also compare whether codon usage biases are similar between genomes using Wilcoxon Two Sample Test. The CodonO webserver provides all of pre-calculated SCUO and GC compositions for each genome, which can be batch downloaded for the users' perference analyses. The standalone CodonO is freely avaiable HERE.
A protein sequence is a string of amino acids, each of which is encoded by three nucleotides. There are twenty amino acids and typically sixty-one genetic codes for these amino acids. For any given protein, two sources of bias in the codon usage are present: 1) amino acid bias, which is due to the non-uniform distribution of amino acids in protein; 2) synonymous codon usage bias, which is the uneven distribution of synonymous codons, i.e., various synonymous codons are not equally used to represent a given amino acid. Within the standard genetic codes, all amino acids except Met and Trp are coded by more than one codon.
DNA sequence data from diverse organisms clearly show that synonymous codons for any amino acid are not used with equal frequency, even though choices among these codons are equivalent in terms of protein sequences (Grantham et al., 1980; Aota and Ikemura, 1986; Murray et al., 1989; Sharp et al., 1988; Shields et al., 1988; D’Onofrio et al., 1991). The relative frequency of synonymous codons varies with both the genes and the organisms. In Escherichia coli and Saccharomyces cerevisiae, codon usage correlates with tRNA content and highly expressed genes frequently use codons corresponding to the most abundant tRNAs (Ikemura, 1985). In contrast, non-coding regions of E. coli DNA showed no pronounced preference for any codon. Recently, the constraints of tRNA contents on synonymous codon choice were confirmed in 18 different unicellular organisms (Kanaya et al., 1999; Rocha et al., 2004). In addition, codon usage bias has been shown to reduce the level of error in translation of the genetic code (Archetti, 2004). In eukaryotes, codon usage bias may be affected by the selection at the pre-mRNA level (Willie and Majewski, 2004). In vertebrates, CpG suppression and DNA methylation effects (Tazi and Bird, 1990), mRNA stability (Holmquist and Filipski, 1994), codon context (Karlin and Mrazek, 1996), and species of origin (Lawrence and Ochman, 1997) have been shown to influence the codon usage bias levels as well (reviewed in Karlin et al., 1998). The codon usage bias was also associated with tissue or organ specificity (Holmquist and Filipski, 1994). However, Zhang and Li (2004) further found that the codon usage pattern of housekeeping genes does not seem to differ from that of tissue-specific genes.
Quantification of codon usage bias helps understand evolution of living organisms and genome analyses. Many different approaches have been developed in the past few decades. Most of these existing computational approaches are only suitable for the comparison of codon usage bias within a single genome. Synonymous Codon Usage Order, SCUO, is a new index developed to measure Synonymous Codon Usage Bias using information theory (Wan et al. 2003, 2004). Different from other methods, SCUO is fit for measuring synonymous codon usage bias within and across genomes. The reviews of the codon usage bias measurement methods are available in Wan et al. (2006).
References
- Aota, S. and Ikemura, T. (1986) Diversity in G + C content at the third position of codons in vertebrate genes and its cause. Nucleic Acids Res., 14, 6345-6355.
- Archetti, M. (2004) Codon usage bias and mutation constraints reduce the level of error minimization of the genetic code. J Mol Evol., 59, 258-266.
- D'Onofrio, G., Mouchiroud, D., Aissani, B., Gautier, C. and Bernardi, G. (1991) Correlations between the compositional properties of human genes, codon usage, and amino acid composition of proteins. J. Mol. Evol., 32, 504-510.
- Grantham, R., Gautier, C. and Gouy, M. (1980) Codon frequencies in 119 individual genes confirm consistent choices of degenerate bases according to genome type. Nucleic Acids Res., 8, 1893-1912.
- Holmquist, G. P. and Filipski, J. (1994) Organization of mutations along the genome: a prime determinant of genome evolution. Trends Ecol. Evol., 9, 65-69.
- Ikemura, T. (1985) Codon usage and tRNA content in unicellular and multicellular organisms. Mol. Biol. Evol., 2, 13-34.
- Lawrence, J.G. and Ochman, H. (1997) Amelioration of bacterial genomes: rates of change and exchange. J. Mol. Evol., 44, 383-397.
- Kanaya, S., Yamada, Y., Kudo, Y. and Ikemura, T. (1999) Studies of codon usage and tRNA genes of 18 unicellular organisms and quantification of Bacillus subtilis tRNAs: gene expression level and species-specific diversity of codon usage based on multivariate analysis. Gene, 238, 143-155.
- Karlin, S. and Mrazek, J. (1996) What drives codon choices in human genes? J. Mol. Biol., 262, 459-472.
- Karlin, S., Campbell, A. M. and Mrazek, J. (1998) Comparative DNA analysis across diverse genomes. Annu. Rev. Genet., 32, 185-225.
- Murray, E. E., Lotzer, J. and Eberle, M. (1989) Codon usage in plant genes. Nucleic Acids Res., 17, 477-498.
- Sharp, P. M., Cowe, E., Higgins, D. G., Shields, D. C., Wolfe, K. H. and Wright, F. (1988) Codon usage patterns in Escherichia coli, Bacillus subtilis, Saccharomyces cerevisiae, Schizosaccharomyces pombe, Drosophila melanogaster and Homo sapiens; a review of the considerable within-species diversity. Nucleic Acids Res., 16, 8207-8211.
- Shields, D. C., Sharp, P. M., Higgins, D. G. and Wright, F. (1988) "Silent" sites in Drosophila genes are not neutral: evidence of selection among synonymous codons. Mol. Biol. Evol., 5, 704-716.
- Tazi, J. and Bird, A. (1993) Alternative chromatin structure at CpG islands. Cell, 60, 909-920.
- Wan, X., D. Xu, and J. Zhou. 2003. A new informatics method measuring synonymous codon usage bias. In Intelligent engineering systems through artificial neural networks, vol 13, eds. Dagli et al., 1101-1018. New York, NY: ASME Press.
- Wan, X.-F., Xu, D., Kleinhofs, A., and Zhou J. (2004) Quantitative relationship between synonymous codon usage bias and GC composition across unicellular genomes. BMC Evol Biol., 4, 19.
- Wan, X.-F., J. Zhou, and D. Xu. 2006. CodonO: a new informatics method measuring synonymous codon usage bias. International Journal of General Systems, 35: 109-125.
- Willie, E. and Majewski, J. (2004) Evidence for codon bias selection at the pre-mRNA level in eukaryotes. Trends Genet., 20, 534-538.
- Zhang, L. and Li, W. H. (2004) Mammalian housekeeping genes evolve more slowly than tissue-specific genes. Mol Biol Evol. 21, 236-239.
Synonymous Codon Usage Order (SCUO)
To implement the informatics method, SCUO, we created a codon table for the amino acids that have more than one codon, indexed in an arbitrary way, so that we may unambiguously refer to the jth (degenerate) codon of amino acid i, 1 ≤ i ≤ 18. In mycoplasmas, Trp was also included into the codon table since a standard stop codon TGA encodes Trp in this specific species so that 1 ≤ i ≤ 19. To simplify the explanation, the following description of the method is only based on the standard genetic codon table although the actual SCUO computation considered special cases for different organisms. Let ![]() represent the number of degenerate codons for amino acid i, so 1 ≤ j ≤
represent the number of degenerate codons for amino acid i, so 1 ≤ j ≤ ![]() ; for example, 1 ≤ j ≤ 6 for leucine, 1 ≤ j ≤ 2 for tyrosine, etc. For each sequence, let
; for example, 1 ≤ j ≤ 6 for leucine, 1 ≤ j ≤ 2 for tyrosine, etc. For each sequence, let ![]() represent the occurrence of synonymous codon j for amino acid i, 1 ≤ i ≤ 18, 1 ≤ j ≤
represent the occurrence of synonymous codon j for amino acid i, 1 ≤ i ≤ 18, 1 ≤ j ≤![]() . Normalizing the
. Normalizing the ![]() by their sum over j gives the frequency of the jth degenerate codon for amino acid i in each sequence
by their sum over j gives the frequency of the jth degenerate codon for amino acid i in each sequence
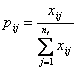
According to information theory, we define the entropy ![]() of the ith amino acid of the jth codon in each sequence by
of the ith amino acid of the jth codon in each sequence by
![]()
Summing over the codons representing amino acid i gives the entropy of the ith amino acid in the each sequence
![]()
If the synonymous codons for the ith amino acid were used at random, one would expect a uniform distribution of them as representatives for the ith amino acid. Thus, the maximum entropy for the ith amino acid in each sequence is
![]()
If only one of the synonymous codons is used for the ith amino acid, i.e., the usage of the synonymous codons is biased to the extreme, then the ith amino acid in each sequence has the minimum entropy:
![]()
This information measures the non-randomness in synonymous codon usage and therefore describes the degree of organization for synonymous codon usage for the ith amino acid in each sequence.
![]()
Let ![]() be the normalized difference between the maximum entropy and the observed entropy for the ith amino acid in each sequence, i.e.
be the normalized difference between the maximum entropy and the observed entropy for the ith amino acid in each sequence, i.e.
![]()
Obviously, 0 ≤ ![]() ≤ 1. When synonymous codon usage for the ith amino acid is random,
≤ 1. When synonymous codon usage for the ith amino acid is random, ![]() = 0. When this usage is biased to the extreme,
= 0. When this usage is biased to the extreme, ![]() = 1. Thus,
= 1. Thus, ![]() can be thought as a measure of the bias in synonymous codon usage for the ith amino acid in each sequence. We designate the statistics
can be thought as a measure of the bias in synonymous codon usage for the ith amino acid in each sequence. We designate the statistics ![]() as the synonymous codon usage order (SCUO) for the ith amino acid in each sequence.
as the synonymous codon usage order (SCUO) for the ith amino acid in each sequence.
Let ![]() be the composition ratio of the ith amino acid in each sequence:
be the composition ratio of the ith amino acid in each sequence:
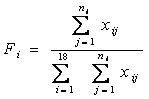
Then the average SCUO for each sequence can be represented as
![]()
The SCUO represents the overall synonymous codon usage order for the sequence.